Generating a non-periodic system for FHI-aims
Objective
Cut out the peptide with a solvation shell of water molecules that can be simulated with FHI-aims without periodic boundary conditions, i.e., as a droplet in vaccuum.
Protocol
The following commands can be used interactively in the python or
ipython interpreter or a Jupyter notebook. Alternatively, combine
them in a script extract_nonpbc.py and run the
script with python extract_nonpbc.py.
Set-up
Import MDAnalysis and the transformations module:
Load the topology (TPR) and trajectory (TRR) of the classical MD
simulation into the universe u:
Change atom names to elements because these names are written to the output file and FHI-aims wants elements, not force field atom names:
Note
With TPR files, information is available for elements. With other input files, MDAnalysis will have to guess the elements.
Create AtomGroup instances for the water molecules (water) and the
capped lysine (solute) by using selection by residue name
We can check that we partitioned the whole system into solute and
water with a simple assertion that the combined AtomGroup of solute +
water is the same as all the atoms in the system:
If the assertion fails then you should check your selections.
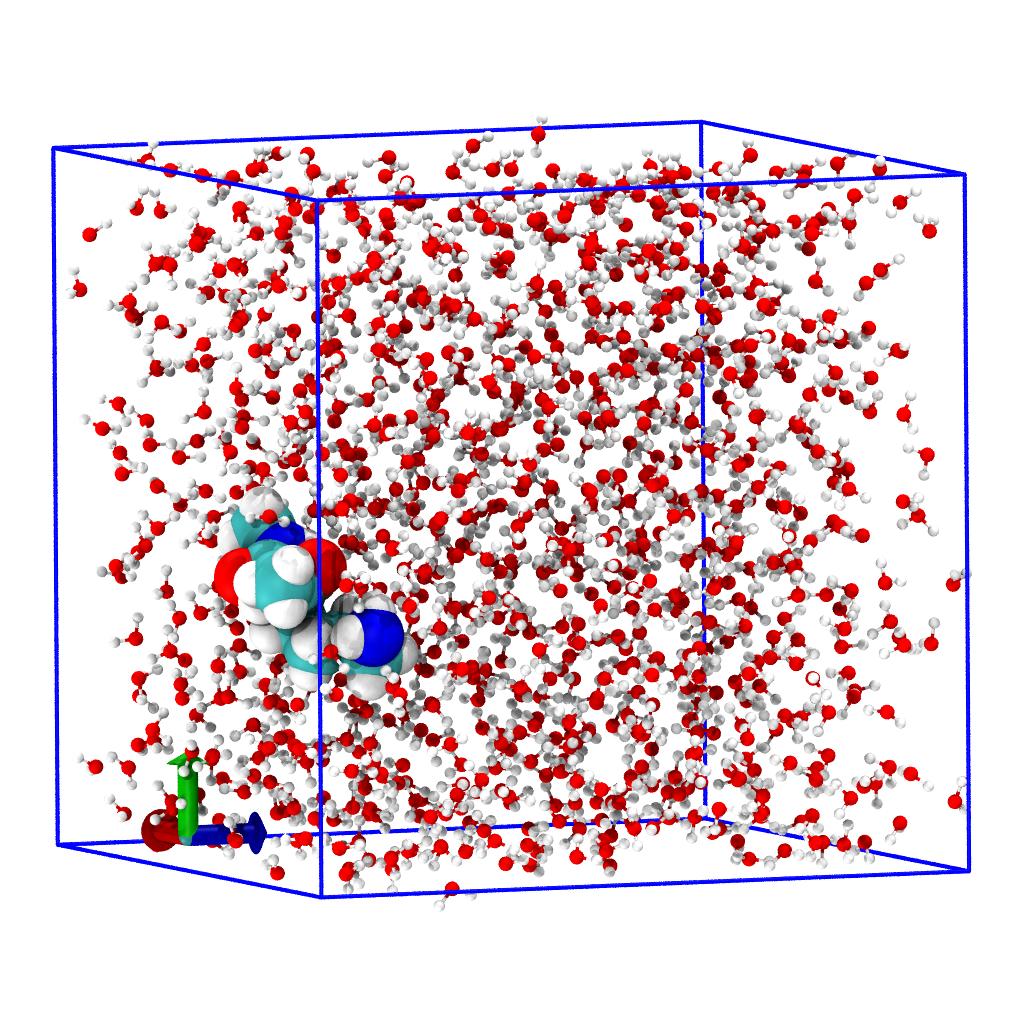
Centering and fixing PBC artifacts
The objective is to generate a trajectory where
- the peptide is at the center of the simulation cell
- water is packed around the peptide
- water molecule are made whole and not broken across periodic boundaries.
We use on-the-fly transformations to rewrite the coordinates of the trajectory while reading the TRR file from the disk. We create a workflow of functions that is run between reading the coordinates from file and storing them in the Timestep instance for the rest of MDAnalysis to see.
workflow = [transformations.unwrap(u.atoms),
transformations.center_in_box(solute, center='geometry'),
transformations.wrap(water, compound='fragments')]
u.trajectory.add_transformations(*workflow)
unwrap(u.atoms)unwraps all molecules over the box boundaries if necessary.center_in_box(solute, center='geometry')shifts all coordinates so that the center of the box coincides with the center of geometry of the AtomGroupsolute(which we selected previously).wrap(water, compound='fragments')ensures that allwatermolecules ("fragments" in the sense of chemically bonded atoms) are in the box that was centered on the solute in transformation 2. (One could also usecompound="residues"in this case for potential speed-ups but "fragments" is the safest choice when real (classical) bonding information is available, as is in the TPR.)
We then add the workflow to the u.trajectory instance and activate
it for all future use of the universe instance.
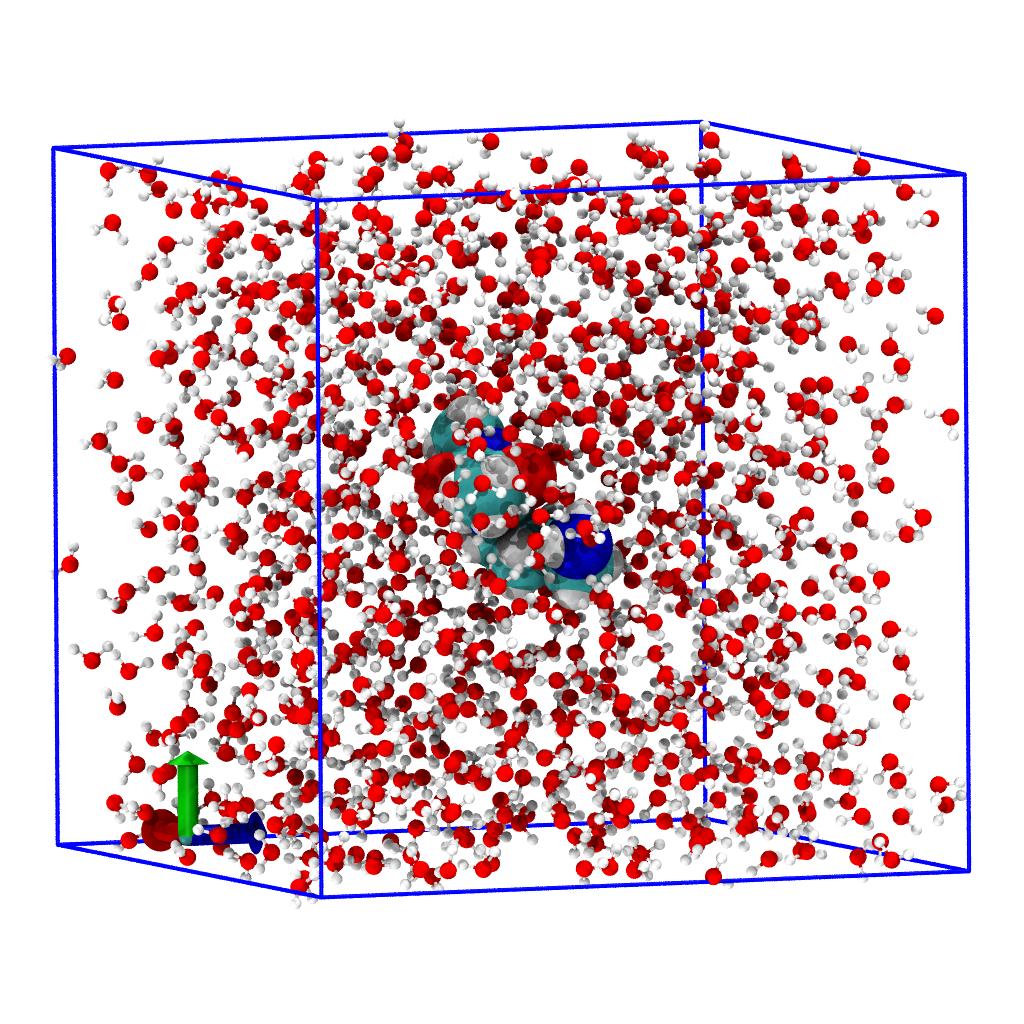
Select trajectory frame
MDAnalysis keeps track of the current frame of the trajectory. At the beginning, the frame "cursor" points to frame 0.
Info
MDAnalysis uses zero-based indexing for frames.
You can move to any frame between 0 and u.trajectory.n_frames - 1 by indexing the u.trajectory
instance, i.e., u.trajectory[frame].
To move to the last frame of trajectory use
All AtomGroups will now read data from the last trajectory frame (suitably transformed).
Subsystem selection
We want to select water molecules within 3 Å of the solute, i.e., the first solvation shell.
We create an AtomGroup named close_water with a selection
command where around 3 <selection> indicates any atom within 3 Å of
the other selection:
Note that we can use our solute AtomGroup inside the selection with
the group <name> selection keyword where <name> is then used as a
Python keyword for select_atoms().
Note
The geometric selection will be applied to the coordinates at the
currently selected trajectory frame. The resulting AtomGroup is
"static" and will not change if the trajectory frame changes. If
a dynamic
AtomGroup
is desired, add the keyword updating=True.
We construct a new group by adding our solute solute and the
close_water and naming it small_system.
Danger
We use the .residues attribute of the AtomGroups in order to include all
atoms in the residue. The original around selection only
included atoms and may well cut water molecules in half. These
incomplete water molecules would present themselves as radicals in
FHI-aims and likely lead to incorrect calculations.
Note
small_system is a ResidueGroup. If we need the corresponding
AtomGroup then we simply use small_system.atoms.
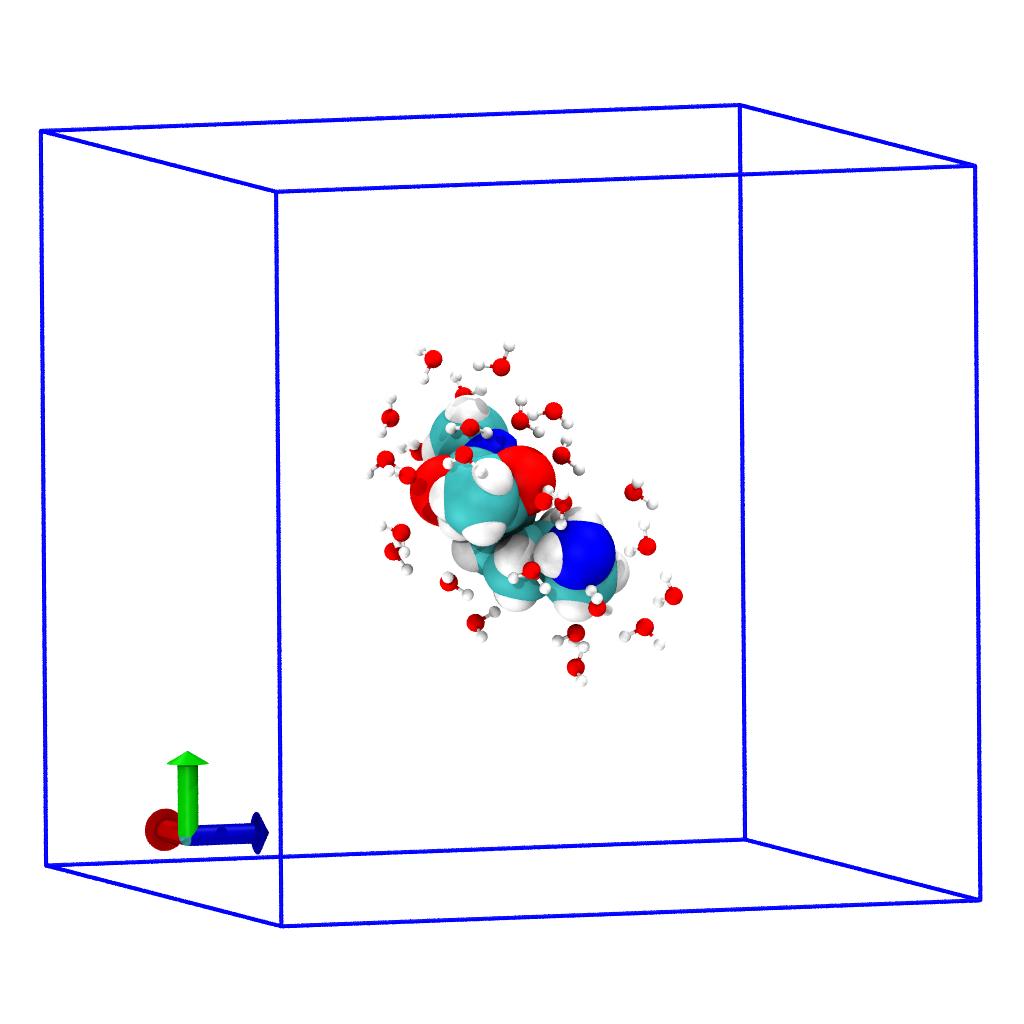
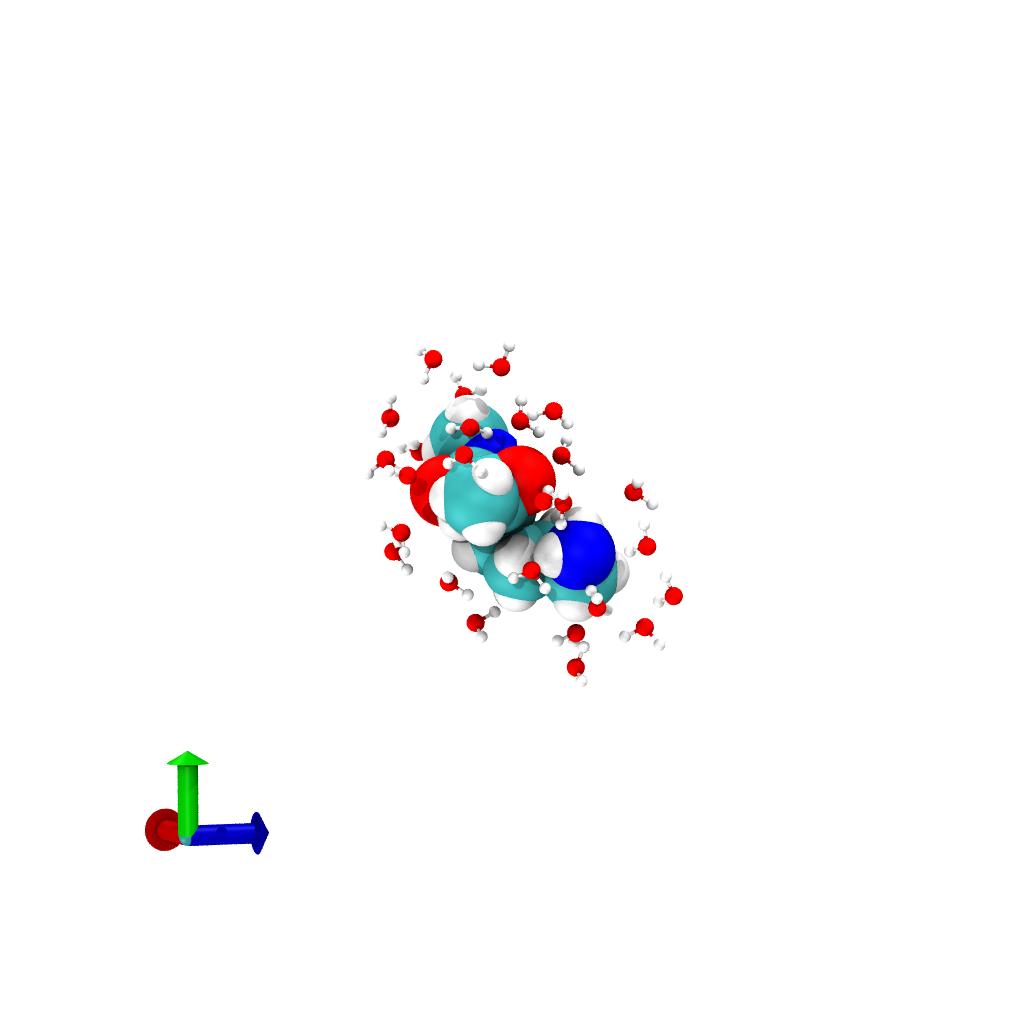
close_water and solute system:
Selection of water (balls and sticks) within 3 Å around the solute
(VDW spheres). The periodic unitcell, which is still present, is
indicated by the blue wireframe box.Remove periodicity
Our objective is to perform non-periodic calculations in
FHI-aims. We therefore do not want the periodic box information from
the classical MD simulation to appear in the geometry.in file.
We can remove periodicity for this write operation with the following hack:
Note
Removing periodicity is not permanent in the MDAnalysis
Universe. The box dimensions will be re-read from the trajectory
file as soon as we move to another frame. However, it is possible
to permanently remove the periodicity using the
boxdimensions transformation.
Output to geometry.in
We want to write the our reduced system to a FHI-aims geometry.in
file (MDAnalysis supports reading and writing the FHI-aims file
format).
We can also write a file in PDB format for visualization in tools that may not understand the FHI-aims format (ignore warnings about attributes that are set to default values to satisfy the PDB standard):
Suppressing warnings in Python
You can temporarily silence the warnings with the warnings filter:
Complete script
Save this script as extract_nonpbc.py:
import MDAnalysis as mda
from MDAnalysis import transformations
u = mda.Universe("./files/MD/NPT/md.tpr", "./files/MD/NPT/md.trr")
u.atoms.names = u.atoms.elements
water = u.atoms.select_atoms("resname SOL")
solute = u.atoms.select_atoms("resname ACE LYSN NME")
assert u.atoms == solute + water
workflow = [transformations.unwrap(u.atoms),
transformations.center_in_box(solute, center='geometry'),
transformations.wrap(water, compound='fragments')]
u.trajectory.add_transformations(*workflow)
u.trajectory[-1]
close_water = u.select_atoms("resname SOL and around 3 group SOLUTE",
SOLUTE=solute)
small_system = solute.residues + close_water.residues
small_system.dimensions = None
small_system.atoms.write("geometry.in")
small_system.atoms.write("geometry.pdb")
Adjust the input files and the selection criteria and then run it with